Tandemers (Metabolic Flux Analysis via MS/MS data)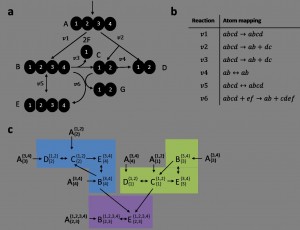
MS/MS enables the measurement of a metabolite tandem mass-isotopomer distribution, representing the abundance in which certain parent and product fragments of a metabolite have different number of labeled atoms. The tandemer approach efficiently computes metabolite tandem mass-isotopomer distributions in a metabolic network, given some estimation of metabolic fluxes, facilitating efficient usage of MS/MS data for metabolic flux analysis [implementation]
Metabolic Flux Analysis via MS/MS data: An efficient method for modeling tandem mass-isotopomer distributions
N. Tepper and T. Shlomi (Submitted)
MFA/UF (Metabolic Flux Analysis/Unknown Fragments)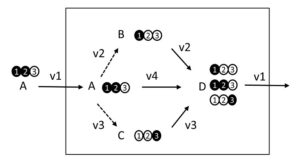
We present a novel method, MFA/UF (Metabolic Flux Analysis/Unknown
Fragments), that can utilize tandem-MS data for flux inference even when the positional origin of collisional fragments is unknown. This is demonstrated by extending the current MFA framework to jointly search for the most likely metabolic flux rates together with the most plausible position of collisional fragments that would optimally match measured tandem-MS data. [implementation]
An integrated computational approach for metabolic flux analysis coupled with inference of tandem-MS collisional fragment positions.
N. Tepper and T. Shlomi
MIRAGE (MetabolIc Reconstruction via functionAl GEnomics)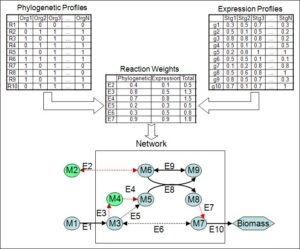
We present a novel gap-filling approach, MetabolIc Reconstruction via functionAl GEnomics (MIRAGE), which identifies missing network reactions
by integrating metabolic flux analysis and functional genomics data.
Specifically, to reconstruct a metabolic network model for an organism of
interest, MIRAGE starts from a core set of reactions, whose presence is
established via strong genomic evidence, and identifies missing reactions
from other species that are required to activate the latter core
reactions, whose presence is further supported by phylogenetic and gene
expression data. [Cyanobacteria models (SBML)] [implementation]
Functional genomics-based approach for metabolic network model
reconstruction: Application for cyanobacteria networks.
E. Vitkin and T. Shlomi